82 Eukaryotic Transcription Gene Regulation
Learning Objectives
By the end of this section, you will be able to do the following:
- Discuss the role of transcription factors in gene regulation
- Explain how enhancers and repressors regulate gene expression
Like prokaryotic cells, the transcription of genes in eukaryotes requires the action of an RNA polymerase to bind to a DNA sequence upstream of a gene in order to initiate transcription. However, unlike prokaryotic cells, the eukaryotic RNA polymerase requires other proteins, or transcription factors, to facilitate transcription initiation. RNA polymerase by itself cannot initiate transcription in eukaryotic cells. There are two types of transcription factors that regulate eukaryotic transcription: General (or basal) transcription factors bind to the core promoter region to assist with the binding of RNA polymerase. Specific transcription factors bind to various regions outside of the core promoter region and interact with the proteins at the core promoter to enhance or repress the activity of the polymerase.
View the process of transcription—the making of RNA from a DNA template.
The Promoter and the Transcription Machinery
Genes are organized to make the control of gene expression easier. The promoter region is immediately upstream of the coding sequence. This region can be short (only a few nucleotides in length) or quite long (hundreds of nucleotides long). The longer the promoter, the more available space for proteins to bind. This also adds more control to the transcription process. The length of the promoter is gene-specific and can differ dramatically between genes. Consequently, the level of control of gene expression can also differ quite dramatically between genes. The purpose of the promoter is to bind transcription factors that control the initiation of transcription.
Within the core promoter region, 25 to 35 bases upstream of the transcriptional start site, resides the TATA box. The TATA box has the consensus sequence of 5’-TATAAA-3’. The TATA box is the binding site for a protein complex called TFIID, which contains a TATA-binding protein. Binding of TFIID recruits other transcription factors, including TFIIB, TFIIE, TFIIF, and TFIIH. Some of these transcription factors help to bind the RNA polymerase to the promoter, and others help to activate the transcription initiation complex.
In addition to the TATA box, other binding sites are found in some promoters. Some biologists prefer to restrict the range of the eukaryotic promoter to the core promoter, or polymerase binding site, and refer to these additional sites as promoter-proximal elements, because they are usually found within a few hundred base pairs upstream of the transcriptional start site. Examples of these elements are the CAAT box, with the consensus sequence 5’-CCAAT-3’ and the GC box, with the consensus sequence 5’-GGGCGG-3’. Specific transcription factors can bind to these promoter-proximal elements to regulate gene transcription. A given gene may have its own combination of these specific transcription-factor binding sites. There are hundreds of transcription factors in a cell, each of which binds specifically to a particular DNA sequence motif. When transcription factors bind to the promoter just upstream of the encoded gene, it is referred to as a cis-acting element, because it is on the same chromosome just next to the gene. Transcription factors respond to environmental stimuli that cause the proteins to find their binding sites and initiate transcription of the gene that is needed.
Enhancers and Transcription
In some eukaryotic genes, there are additional regions that help increase or enhance transcription. These regions, called enhancers, are not necessarily close to the genes they enhance. They can be located upstream of a gene, within the coding region of the gene, downstream of a gene, or may be thousands of nucleotides away.
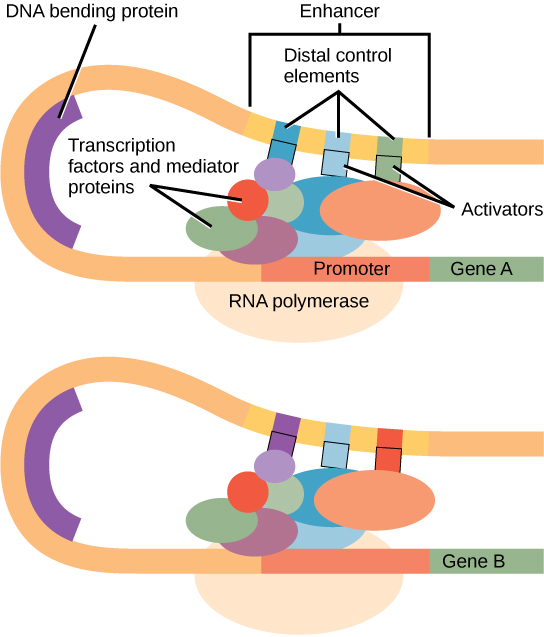
Enhancer regions are binding sequences, or sites, for specific transcription factors. When a protein transcription factor binds to its enhancer sequence, the shape of the protein changes, allowing it to interact with proteins at the promotor site. However, since the enhancer region may be distant from the promoter, the DNA must bend to allow the proteins at the two sites to come into contact. DNA bending proteins help to bend the DNA and bring the enhancer and promoter regions together ((Figure)). This shape change allows for the interaction of the specific activator proteins bound to the enhancers with the general transcription factors bound to the promoter region and the RNA polymerase.
Turning Genes Off: Transcriptional Repressors
Like prokaryotic cells, eukaryotic cells also have mechanisms to prevent transcription. Transcriptional repressors can bind to promoter or enhancer regions and block transcription. Like the transcriptional activators, repressors respond to external stimuli to prevent the binding of activating transcription factors.
Section Summary
To start transcription, general transcription factors, such as TFIID, TFIIB, and others, must first bind to the TATA box and recruit RNA polymerase to that location. Additional transcription factors may also bind to other regulatory elements at the promoter to increase or prevent transcription. In addition to promoter sequences, enhancer regions help augment transcription. Enhancers can be upstream, downstream, within a gene itself, or on other chromosomes. Specific transcription factors bound to enhancer regions may either increase or prevent transcription.
Review Questions
The binding of ________ is required for transcription to start.
- a protein
- DNA polymerase
- RNA polymerase
- a transcription factor
C
What will result from the binding of a transcription factor to an enhancer region?
- decreased transcription of an adjacent gene
- increased transcription of a distant gene
- alteration of the translation of an adjacent gene
- initiation of the recruitment of RNA polymerase
B
A scientist compares the promoter regions of two genes. Gene A’s core promoter plus proximal promoter elements encompasses 70bp. Gene B’s core promoter plus proximal promoter elements encompasses 250bp. Which of the scientist’s hypotheses is most likely to be correct?
- More transcripts will be made from Gene B.
- Transcription of Gene A involves fewer transcription factors.
- Enhancers control Gene B’s transcription.
- Transcription of Gene A is more controlled than transcription of Gene B.
B
Critical Thinking Questions
A mutation within the promoter region can alter transcription of a gene. Describe how this can happen.
A mutation in the promoter region can change the binding site for a transcription factor that normally binds to increase transcription. The mutation could either decrease the ability of the transcription factor to bind, thereby decreasing transcription, or it can increase the ability of the transcription factor to bind, thus increasing transcription.
What could happen if a cell had too much of an activating transcription factor present?
If too much of an activating transcription factor were present, then transcription would be increased in the cell. This could lead to dramatic alterations in cell function.
A scientist identifies a potential transcription regulation site 300bp downstream of a gene and hypothesizes that it is a repressor. What experiment (with results) could he perform to support this hypothesis?
The easiest way to test his hypothesis would be to mutate the site in a cell, and monitor levels of the mRNA transcript made from the gene. If the levels of transcript increase in the mutated cell, then the site was repressing transcription.
Glossary
- cis-acting element
- transcription factor binding sites within the promoter that regulate the transcription of a gene adjacent to it
- enhancer
- segment of DNA that is upstream, downstream, perhaps thousands of nucleotides away, or on another chromosome that influence the transcription of a specific gene
- trans-acting element
- transcription factor binding site found outside the promoter or on another chromosome that influences the transcription of a particular gene
- transcription factor binding site
- sequence of DNA to which a transcription factor binds

