81 Eukaryotic Epigenetic Gene Regulation
Learning Objectives
By the end of this section, you will be able to do the following:
- Explain how chromatin remodeling controls transcriptional access
- Describe how access to DNA is controlled by histone modification
- Describe how DNA methylation is related to epigenetic gene changes
Eukaryotic gene expression is more complex than prokaryotic gene expression because the processes of transcription and translation are physically separated. Unlike prokaryotic cells, eukaryotic cells can regulate gene expression at many different levels. Epigenetic changes are inheritable changes in gene expression that do not result from changes in the DNA sequence. Eukaryotic gene expression begins with control of access to the DNA. Transcriptional access to the DNA can be controlled in two general ways: chromatin remodeling and DNA methylation. Chromatin remodeling changes the way that DNA is associated with chromosomal histones. DNA methylation is associated with developmental changes and gene silencing.
Epigenetic Control: Regulating Access to Genes within the Chromosome
The human genome encodes over 20,000 genes, with hundreds to thousands of genes on each of the 23 human chromosomes. The DNA in the nucleus is precisely wound, folded, and compacted into chromosomes so that it will fit into the nucleus. It is also organized so that specific segments can be accessed as needed by a specific cell type.
The first level of organization, or packing, is the winding of DNA strands around histone proteins. Histones package and order DNA into structural units called nucleosome complexes, which can control the access of proteins to the DNA regions ((Figure)a). Under the electron microscope, this winding of DNA around histone proteins to form nucleosomes looks like small beads on a string ((Figure)b).
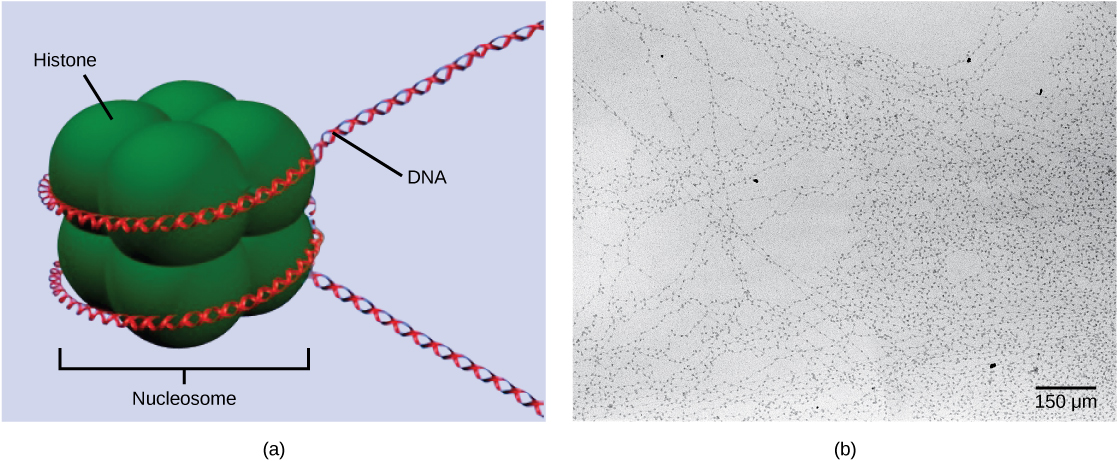
These beads (histone proteins) can move along the string (DNA) to expose different sections of the molecule. If DNA encoding a specific gene is to be transcribed into RNA, the nucleosomes surrounding that region of DNA can slide down the DNA to open that specific chromosomal region and allow for the transcriptional machinery (RNA polymerase) to initiate transcription ((Figure)).
In females, one of the two X chromosomes is inactivated during embryonic development because of epigenetic changes to the chromatin. What impact do you think these changes would have on nucleosome packing?
<!– <link window=”new” target-id=”fig-ch16_03_02″ document=””/> The nucleosomes would pack more tightly together. –>
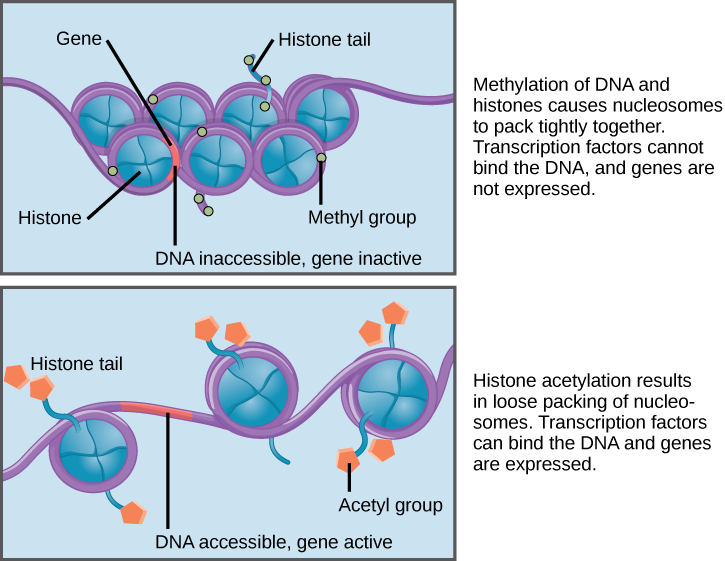
How closely the histone proteins associate with the DNA is regulated by signals found on both the histone proteins and on the DNA. These signals are functional groups added to histone proteins or to DNA and determine whether a chromosomal region should be open or closed ((Figure) depicts modifications to histone proteins and DNA). These tags are not permanent, but may be added or removed as needed. Some chemical groups (phosphate, methyl, or acetyl groups) are attached to specific amino acids in histone “tails” at the N-terminus of the protein. These groups do not alter the DNA base sequence, but they do alter how tightly wound the DNA is around the histone proteins. DNA is a negatively charged molecule and unmodified histones are positively charged; therefore, changes in the charge of the histone will change how tightly wound the DNA molecule will be. By adding chemical modifications like acetyl groups, the charge becomes less positive, and the binding of DNA to the histones is relaxed. Altering the location of nucleosomes and the tightness of histone binding opens some regions of chromatin to transcription and closes others.
The DNA molecule itself can also be modified by methylation. DNA methylation occurs within very specific regions called CpG islands. These are stretches with a high frequency of cytosine and guanine dinucleotide DNA pairs (CG) found in the promoter regions of genes. The cytosine member of the CG pair can be methylated (a methyl group is added). Methylated genes are usually silenced, although methylation may have other regulatory effects. In some cases, genes that are silenced during the development of the gametes of one parent are transmitted in their silenced condition to the offspring. Such genes are said to be imprinted. Parental diet or other environmental conditions may also affect the methylation patterns of genes, which in turn modifies gene expression. Changes in chromatin organization interact with DNA methylation. DNA methyltransferases appear to be attracted to chromatin regions with specific histone modifications. Highly methylated (hypermethylated) DNA regions with deacetylated histones are tightly coiled and transcriptionally inactive.
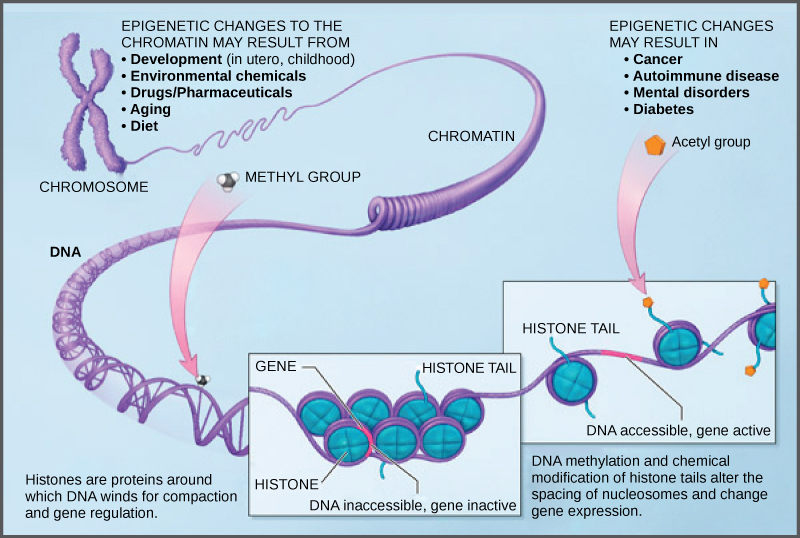
Epigenetic changes are not permanent, although they often persist through multiple rounds of cell division and may even cross generational lines. Chromatin remodeling alters the chromosomal structure (open or closed) as needed. If a gene is to be transcribed, the histone proteins and DNA in the chromosomal region encoding that gene are modified in a way that opens the promoter region to allow RNA polymerase and other proteins, called transcription factors, to bind and initiate transcription. If a gene is to remain turned off, or silenced, the histone proteins and DNA have different modifications that signal a closed chromosomal configuration. In this closed configuration, the RNA polymerase and transcription factors do not have access to the DNA and transcription cannot occur ((Figure)).
View this video that describes how epigenetic regulation controls gene expression.
Section Summary
In eukaryotic cells, the first stage of gene-expression control occurs at the epigenetic level. Epigenetic mechanisms control access to the chromosomal region to allow genes to be turned on or off. Chromatin remodeling controls how DNA is packed into the nucleus by regulating how tightly the DNA is wound around histone proteins. The DNA itself may be methylated to selectively silence genes. The addition or removal of chemical modifications (or flags) to histone proteins or DNA signals the cell to open or close a chromosomal region. Therefore, eukaryotic cells can control whether a gene is expressed by controlling accessibility to the binding of RNA polymerase and its transcription factors.
Visual Connection Questions
Review Questions
What are epigenetic modifications?
- the addition of reversible changes to histone proteins and DNA
- the removal of nucleosomes from the DNA
- the addition of more nucleosomes to the DNA
- mutation of the DNA sequence
A
Which of the following are true of epigenetic changes?
- allow DNA to be transcribed
- move histones to open or close a chromosomal region
- are temporary
- all of the above
D
Critical Thinking Questions
In cancer cells, alteration to epigenetic modifications turns off genes that are normally expressed. Hypothetically, how could you reverse this process to turn these genes back on?
You can create medications that reverse the epigenetic processes (to add histone acetylation marks or to remove DNA methylation) and create an open chromosomal configuration.
A scientific study demonstrated that rat mothering behavior impacts the stress response in their pups. Rats that were born and grew up with attentive mothers showed low activation of stress-response genes later in life, while rats with inattentive mothers had high activation of stress-response genes in the same situation. An additional study that swapped the pups at birth (i.e., rats born to inattentive mothers grew up with attentive mothers and vice versa) showed the same positive effect of attentive mothering. How do genetics and/or epigenetics explain the results of this study?
Swapping the pups at birth indicates that the genes inherited from the attentive or inattentive mothers do not explain the rats’ stress-responses later in life. Instead, researchers found that the attentive mothering caused the methylation of genes that control the expression of stress receptors in the brain. Thus, rats that received attentive maternal care exhibited epigenetic changes that limited the expression of stress-response genes, and that the effect was durable over their lifespans.
Some autoimmune diseases show a positive correlation with dramatically decreased expression of histone deacetylase 9 (HDAC9, an enzyme that removes acetyl groups from histones). Why would the decreased expression of HDAC9 cause immune cells to produce inflammatory genes at inappropriate times?
Histone acetylation reduces the positive charge of histone proteins, loosening the DNA wrapped around the histones. This looser DNA can then interact with transcription factors to express genes found in that region. Normally, once the gene is no longer needed, histone deacetylase enzymes remove the acetyl groups from histones so that the DNA becomes tightly wound and inaccessible again. However, when there is a defect in HDAC9, the deacetylation may not occur. In an immune cell, this would mean that inflammatory genes that were made accessible during an infection are not tightly rewound around the histones.
Glossary
- transcription factor
- protein that binds to the DNA at the promoter or enhancer region and that influences transcription of a gene

